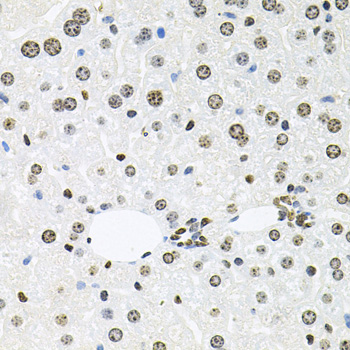
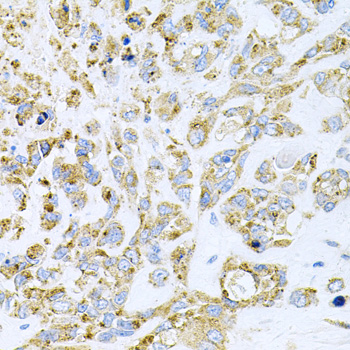
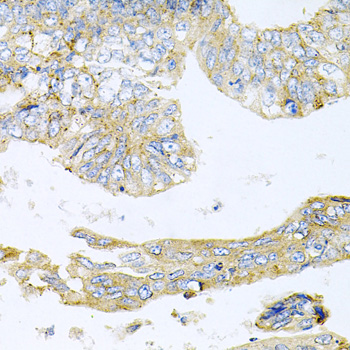
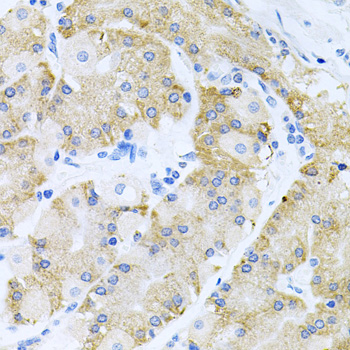
Acetylation of the histone tail causes chromatin to adopt an "open" conformation, allowing increased accessibility of transcription factors to DNA. The identification of histone acetyltransferases (HATs) and their large multiprotein complexes has yielded important insights into how these enzymes regulate transcription (1,2). HAT complexes interact with sequence-specific activator proteins to target specific genes. In addition to histones, HATs can acetylate nonhistone proteins, suggesting multiple roles for these enzymes (3). In contrast, histone deacetylation promotes a "closed" chromatin conformation and typically leads to repression of gene activity (4). Mammalian histone deacetylases can be divided into three classes on the basis of their similarity to various yeast deacetylases (5). Class I proteins (HDACs 1, 2, 3, and 8) are related to the yeast Rpd3-like proteins, those in class II (HDACs 4, 5, 6, 7, 9, and 10) are related to yeast Hda1-like proteins, and class III proteins are related to the yeast protein Sir2. Inhibitors of HDAC activity are now being explored as potential therapeutic cancer agents (6,7).
HDAC3 is a nuclear and cytoplasmic protein that deacetylates both histone (H2A, H3, H4) and non-histone substrates (RelA, SRY, p53, MEF2, PCAF and p300/CBP) (8). HDAC3 deacetylase activity is stimulated by interactions with the N-CoR and SMRT co-repressor proteins. Together, these three proteins form a functional complex that represses transcription associated with nuclear hormone receptors and other transcription factors, including Rev-Erb, COUP-TF, DAX1, MAD and Pit-1 (8,9). Phosphorylation of HDAC3 on Ser424 by casein kinase 2 (CK2) also increases HDAC3 deacetylase activity (9). Subsequently, de-phosphorylation by protein phosphatase 4 (PP4) decreases HDAC3 activity (9).